Structural studies of molecular motors: Relationship with degenerative deseases
Molecular motors are multi-protein complexes that produce important mechanical or chemical work using either adenosine triphosphate (ATP), the energy fuel of the cell, or producing it.
The laboratory is interested in analyzing two splendid molecular motors through structural studies;
- Human dynein is a multi-protein complex with a M. W. of around 1,300 kDa and functions as a molecular motor. This engine is divided into two domains (Figure 1); The "motor" that is the domain that uses ATP to "walk" sequentially to the minus-end of microtubules, and; The "stem" where the dimerization domain is located and where the other subunits are bound. Dynein is composed of two copies of 6 different proteins. This molecular motor, together with its cofactor, dynactin, is responsible for the polarized spatial organization of intra-cellular transport towards the nucleus and also assist in cell division. On the other hand, the medical importance of dynein is that this machine is recruited for viral transport to the nucleus of the cell. In addition, mutations in this complex cause severe neurodegenerative disorders in patients. In this project, fragments of the human dynein and mutants will be purified to solve their crystallographic structures at high resolution and thus to propose the binding mechanism of this molecular motor with its different cargo-cargo adaptors.
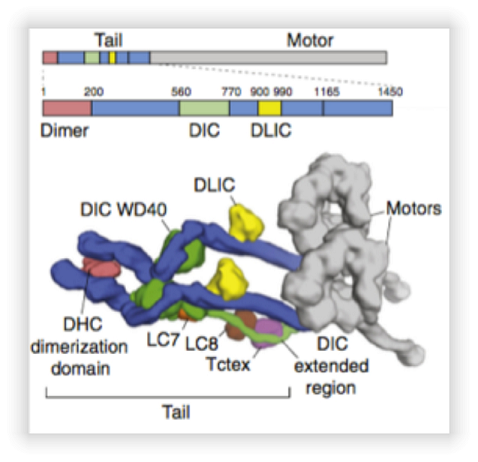
Model of human dynein derived from cryo-electron microscopy. Taken from (Carter, AP, et al., 2016). The top panel shows the gene topology from dynein subunits. The lower panel shows the approximate location of the subunits and different domains of dynein.
- ATP synthase is also a protein complex found in energy transducer membranes; bacterial plasmatic membranes, in plant chloroplast lamellae and in the inner membrane of eukaryotic mitochondria, the molecular weight its among 500 kDa and 850 kDa depending on the species. ATP synthase synthesizes ATP from ADP and Pi using proton motive force as the energy source. The enzyme can be separated into two sectors: The F1 sector, which is water soluble and is where catalytic sites are located; and the hydrophobic Fo sector, embedded in the membrane, here is the channel through which protons pass and produces the rotational movement of the enzyme. Thanks to the latest studies carried out in the ATP synthase from Paracoccus denitrificans has managed to solve a question that had remained in the field for the past 30 years, the crystallographic structure of the complete enzyme (Figure 2). With this structure we have a better understanding of the proton channel mechanism and its coupling to the ATP synthesis. One of the subunits that had remained elusive, the "a" subunit, lies on the membrane at 30o to the central axis of the "c" subunit ring (Figure 3). Mutations in this subunit generate some degenerative diseases. In this project we will solve the atomic structure of ATP synthase with the different mutations in the subunit a, together with biochemical techniques, to determine the molecular mechanism that provokes these mutations in the enzyme.
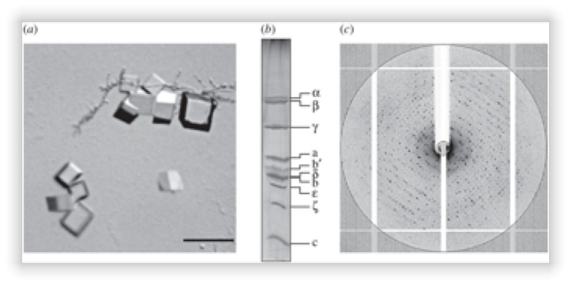
Figure 2. Crystallization and diffraction of ATP synthase from P. denitrificans.
(a) Crystals of the enzyme, the scale measures 50 μm. (b) Polyacrylamide gel denatured in the presence of SDS of dissolved crystals. (c) Diffraction image of a representative crystal.
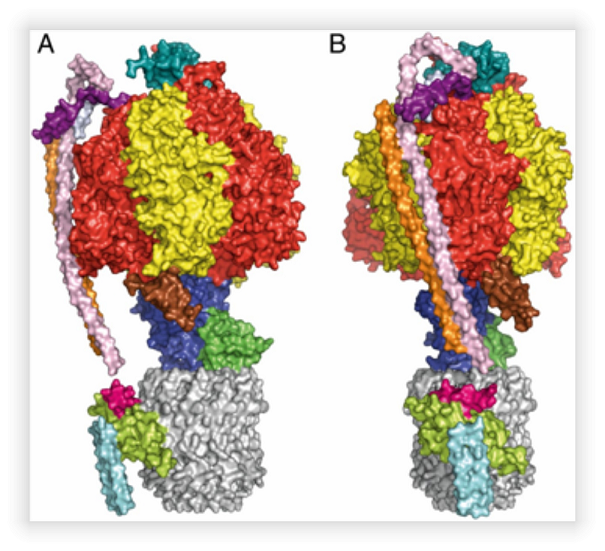
Model derived from diffraction of the crystals of the ATP synthase from P. denitrificans. A. Side view of the complex. B. Frontal view of the enzyme.